The pandas Series is a numpy array with the addition of an index for each entry. It is also the building block object for the pandas DataFrame, like below.
Since the Series is constructed from numpy arrays, this gives them the added benefit of faster computation on a Series or between multiple ones due to numpy's vectorized computation. While with a base python list a loop must be used to perform computation on elements of a list, slowly checking the index and confirming the data type, numpy handles this in pre-compiled C code for much faster computation.
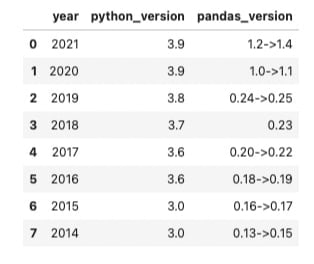
Let's construct the above DataFrame by building a Series for each column and exploring how to change the data type of our values. Although the length of these columns are trivially small, knowing how to maximize the efficiency of your code by choosing the right data type will become important when dealing with larger amounts of data and performing more complex computations.
We'll start by creating a Series for the year column. We can do this by initializing a variable named 'years' that contains a base python list filled with the year for the past 8 years. Then we will pandas Series method. We will also give the Series a name by passing a string to the name = keyword argument. This Series name will also provide pandas with the column name for these values when added to a DataFrame.
import pandas as pd
import numpy as np
years = [2021, 2020, 2019, 2018, 2017, 2016, 2015, 2014]
year_series = pd.Series(data=years, name='year')
year_series
[output]
0 2021
1 2020
2 2019
3 2018
4 2017
5 2016
6 2015
7 2014
Name: year, dtype: int64
As you can see, pandas has automatically generated a numeric index for our Series and decided to use the 'int64' data type for our values based on the data we passed through. Let's use some attributes and methods to explore what this Series is made of.
The .values attribute gives us the list of years we gave to pandas initially, but we see here that it has been turned it into a numpy array.
print(type(year_series.values))
year_series.values
[output]
<class 'numpy.ndarray'>
array([2021, 2020, 2019, 2018, 2017, 2016, 2015, 2014])
Now let's check how much memory are Series is using with the .memory_usage() method, which outputs the amount of space being used in bytes. For a refresher, every byte is 8 bits. Since our Series contains 8 values, the sum of the bytes used for our array will equal the amount of bits used for each element in the array.
Since our data type is 'int64', measured in bits, and we have 8 values, we could assume our Series consumes 64 bits x 8 values = 512 bits. Converting that to bytes, the .memory_usage() method should give us an output of 64bytes.
print(year_series.memory_usage(), 'bytes')
[output]
192 bytes
Interesting, it did not. Let's dig around to find where the other 128 bytes are coming from. The .nbytes attribute will give us the memory consumption of a numpy array. Let's combine it with the .values and .index attributes on the Series object to check the consumption of both.
print(
f'''
values consumption: {year_series.values.nbytes} bytes
index consumption: {year_series.index.nbytes} bytes
''')
[output]
values consumption: 64 bytes
index consumption: 128 bytes
The index is storing its values in twice the amount of memory as the actual data! This is an even more odd behavior when considering the results of checking the dtype of the index, which says the index is using 'int64'.
year_series.index.dtype
[output]
dtype('int64')
This will be the same index behavior when building a DataFrame with the pd.DatFrame() method. To reduce the amount of memory used by the index, let's try creating our own index with the pandas .Index() method. Then we can recreate our Series with our own custom index by passing it to the index= keyword argument. For even more memory efficiency we can alter the dtype used for the underlying data in our Series. Pandas decided to use 'int64' which can store values between -9,223,372,036,854,775,808 and 9,223,372,036,854,775,807. This is far more space than we need. Let's downgrade the data type of our values to 'uint16'. The 'u' is for unsigned, which works well for values that will not be negative like a year and 'int16' for 16 bits, which can store numbers ranging between 0 and 65,535. This is plenty for representing the years of python and the pandas library versions.
index = pd.Index(list(range(len(years))))
year_series2 = pd.Series(years, index=index, dtype='uint16', name='year')
year_series2
[output]
0 2021
1 2020
2 2019
3 2018
4 2017
5 2016
6 2015
7 2014
Name: year, dtype: uint16
Our custom index using the listed range of the length of our data seems to give us the same index as before. Let's see if its memory consumption is the same.
print(
f'''
total consumption: {year_series2.memory_usage()} bytes
values consumption: {year_series2.values.nbytes} bytes
index consumption: {year_series2.index.nbytes} bytes
''')
[output]
total consumption: 80 bytes
values consumption: 16 bytes
index consumption: 64 bytes
Great, that cut the memory consumption of our index in half from 128 bytes to 64 bytes and of our values from 64 bytes to 16 bytes.
Now that we can create a Series and alter the data types used, let's continue building the DatFrame above with a Series for the python version column for each year in our first Series. We already have a custom index created so we will just have to pass that into the new Series. As for the underlying data, since our version numbers are floating point values, our dtype for this Series will default to 'float64' like our Series containing years defaulted to 'int64'. Let's go ahead and downgrade this to 'float32'.
python_versions = [3.9, 3.9, 3.8, 3.7, 3.6, 3.6, 3, 3]
python_series = pd.Series(python_versions, index=index, dtype='float32', name='python_version')
python_series
[output]
0 3.9
1 3.9
2 3.8
3 3.7
4 3.6
5 3.6
6 3.0
7 3.0
Name: python_version, dtype: float32
Now let's check the memory consumption.
print(
f'''
total consumption: {python_series.memory_usage()} bytes
values consumption: {python_series.values.nbytes} bytes
index consumption: {python_series.index.nbytes} bytes
''')
[output]
total consumption: 96 bytes
values consumption: 32 bytes
index consumption: 64 bytes
Not bad, this column will take up more memory than the one with our years, but we were able to decrease the underlying datas memory consumption from 64 bytes to 32 bytes.
Now let's create our last Series that contains the versions of pandas for the years in our first Series. Since each year has multiple pandas updates, these are stored as a sting representing the range of version updates for that year.
pandas_versions = ['1.2->1.4', '1.0->1.1', '0.24->0.25', '0.23', '0.20->0.22',
'0.18->0.19','0.16->0.17','0.13->0.15']
pandas_series = pd.Series(pandas_versions, index=index, name='pandas_version')
pandas_series
[output]
0 1.2->1.4
1 1.0->1.1
2 0.24->0.25
3 0.23
4 0.20->0.22
5 0.18->0.19
6 0.16->0.17
7 0.13->0.15
Name: pandas_version, dtype: object
We can see a string is represented in the Series as the numpy 'object' dtype.
Now for the memory consumption.
print(
f'''
total consumption: {pandas_series.memory_usage()} bytes
values consumption: {pandas_series.values.nbytes} bytes
index consumption: {pandas_series.index.nbytes} bytes
''')
[output]
total consumption: 128 bytes
values consumption: 64 bytes
index consumption: 64 bytes
We can see the 'object' dtype totals to 64bytes for the column, or 8bytes/64bits, per element. Unlike for numeric values, there is not a smaller 'object' dtype to downgrade to.
Now let's finally put these in a DataFrame with the pd.concat() method.
df = pd.concat([year_series2, python_series, pandas_series], axis = 1)
df
[output]
Memory consumption of a this DataFrame can be checked with the same .memory_usage() method as the Series' used.
print(df.memory_usage())
print()
print(f'total memory consumption: {df.memory_usage().sum()} bytes')
[output]
Index 64
year 16
python_version 32
pandas_version 64
dtype: int64
total memory consumption: 176 bytes
For comparison, I created this same DataFrame by passing a dictionary with the values to the pd.DataFrame() constructor method and checked the memory consumption.
data = {'year': years, 'python_versions': python_versions, 'pandas_versions': pandas_versions}
df2 = pd.DataFrame(data=data)
df2
[output]
Memory consumption for this DataFrame.
print(df2.memory_usage())
print()
print(f'total memory consumption: {df2.memory_usage().sum()} bytes')
[output]
Index 128
year 64
python_versions 64
pandas_versions 64
dtype: int64
total memory consumption: 320 bytes
Our DataFrame made from customized Series' added up 176 bytes, while our DataFrame made from the the pd.DatFrame() method and no customizing came in at 320 bytes.That's a 45% increase in memory consumption!
As you can see, changing the data types used in your pandas DataFrames can drastically decrease the amount of memory used. For larger datasets this can have useful effects in both file size and time for computation.








Top comments (0)