What will be scraped
From Organic results:
📌Note: There's a limit of 100 pages on Google Scholar so whenever you see About xxx.xxx results it does not mean that all of these results will be displayed, the same as it on Google Search.
From Cite results:
Prerequisites
Prepare virtual environment and install libraries
If you're on Linux:
python -m venv env && source env/bin/activate
If you're on Windows and using Git Bash:
python -m venv env && source env/Scripts/activate
If you didn't work with a virtual environment before, have a look at the dedicated Python Virtual Environments tutorial using Virtualenv and Poetry blog post of mine to get familiar.
In short, it's a thing that creates an independent set of installed libraries inside different folder including different Python versions that can coexist with each other on the same system thus preventing libraries and Python version conflicts.
📌Note: using virtual environment is not a strict requirement.
Install libraries:
pip install google-search-results pandas
Process
If you don't need an explanation:
Scrape Google Scholar Organic Results using Pagination
import os
from serpapi import GoogleSearch
from urllib.parse import urlsplit, parse_qsl
def organic_results():
print("extracting organic results..")
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar",
"q": "minecraft redstone system structure characteristics strength", # search query
"hl": "en", # language
"as_ylo": "2017", # from 2017
"as_yhi": "2021", # to 2021
"start": "0" # first page
}
search = GoogleSearch(params)
organic_results_data = []
loop_is_true = True
while loop_is_true:
results = search.get_dict()
print(f"Currently extracting page №{results['serpapi_pagination']['current']}..")
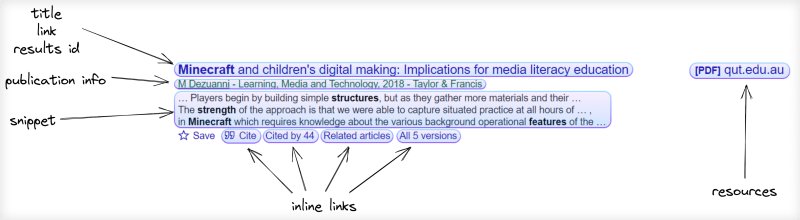
for result in results["organic_results"]:
position = result["position"]
title = result["title"]
publication_info_summary = result["publication_info"]["summary"]
result_id = result["result_id"]
link = result.get("link")
result_type = result.get("type")
snippet = result.get("snippet")
try:
file_title = result["resources"][0]["title"]
except: file_title = None
try:
file_link = result["resources"][0]["link"]
except: file_link = None
try:
file_format = result["resources"][0]["file_format"]
except: file_format = None
try:
cited_by_count = int(result["inline_links"]["cited_by"]["total"])
except: cited_by_count = None
cited_by_id = result.get("inline_links", {}).get("cited_by", {}).get("cites_id", {})
cited_by_link = result.get("inline_links", {}).get("cited_by", {}).get("link", {})
try:
total_versions = int(result["inline_links"]["versions"]["total"])
except: total_versions = None
all_versions_link = result.get("inline_links", {}).get("versions", {}).get("link", {})
all_versions_id = result.get("inline_links", {}).get("versions", {}).get("cluster_id", {})
organic_results_data.append({
"page_number": results["serpapi_pagination"]["current"],
"position": position + 1,
"result_type": result_type,
"title": title,
"link": link,
"result_id": result_id,
"publication_info_summary": publication_info_summary,
"snippet": snippet,
"cited_by_count": cited_by_count,
"cited_by_link": cited_by_link,
"cited_by_id": cited_by_id,
"total_versions": total_versions,
"all_versions_link": all_versions_link,
"all_versions_id": all_versions_id,
"file_format": file_format,
"file_title": file_title,
"file_link": file_link,
})
if "next" in results.get("serpapi_pagination", {}):
search.params_dict.update(dict(parse_qsl(urlsplit(results["serpapi_pagination"]["next"]).query)))
else:
loop_is_true = False
return organic_results_data
Explanation about paginated organic results extraction
Import os, serpapi, urllib libraries:
import os
from serpapi import GoogleSearch
from urllib.parse import urlsplit, parse_qsl
Create and pass search parameters to GoogleSearch() where all extraction happens on the SerpApi backend:
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar",
"q": "minecraft redstone system structure characteristics strength", # search query
"hl": "en", # language
"as_ylo": "2017", # from 2017
"as_yhi": "2021", # to 2021
"start": "0"
}
search = GoogleSearch(params) # extraction happens here
Create temporary list() to store the data that later will be used to save to CSV file or passed to cite_results() function:
organic_results_data = []
Set up a while loop to extract the data from all available pages:
loop_is_true = True
while loop_is_true:
results = search.get_dict()
# data extraction code..
if "next" in results.get("serpapi_pagination", {}):
search.params_dict.update(dict(parse_qsl(urlsplit(results["serpapi_pagination"]["next"]).query)))
else:
loop_is_true = False
- If there's no
"next"page URL available it will break out of thewhileloop by setting aloop_is_truetoFalse. - if there's a
"next"page URL,search.params_dict.updatewill split and update URL toGoogleSearch(params)for a new page.
Extract data in a for loop:
for result in results["organic_results"]:
position = result["position"]
title = result["title"]
publication_info_summary = result["publication_info"]["summary"]
result_id = result["result_id"]
link = result.get("link")
result_type = result.get("type")
snippet = result.get("snippet")
try:
file_title = result["resources"][0]["title"]
except: file_title = None
try:
file_link = result["resources"][0]["link"]
except: file_link = None
try:
file_format = result["resources"][0]["file_format"]
except: file_format = None
try:
cited_by_count = int(result["inline_links"]["cited_by"]["total"])
except: cited_by_count = None
cited_by_id = result.get("inline_links", {}).get("cited_by", {}).get("cites_id", {})
cited_by_link = result.get("inline_links", {}).get("cited_by", {}).get("link", {})
try:
total_versions = int(result["inline_links"]["versions"]["total"])
except: total_versions = None
all_versions_link = result.get("inline_links", {}).get("versions", {}).get("link", {})
all_versions_id = result.get("inline_links", {}).get("versions", {}).get("cluster_id", {})
-
try/exceptwas used to handleNonevalues when they're not returned from Google backend.
If you try to merge everything in one try block, extracted data will be inaccurate meaning if link or snippet is actually present it will return None sometimes instead, that's why there's a lot of try/except blocks.
Append extracted data to temporary list():
organic_results_data = []
# data extraction and a while loop code...
organic_results_data.append({
"page_number": results["serpapi_pagination"]["current"],
"position": position + 1,
"result_type": result_type,
"title": title,
"link": link,
"result_id": result_id,
"publication_info_summary": publication_info_summary,
"snippet": snippet,
"cited_by_count": cited_by_count,
"cited_by_link": cited_by_link,
"cited_by_id": cited_by_id,
"total_versions": total_versions,
"all_versions_link": all_versions_link,
"all_versions_id": all_versions_id,
"file_format": file_format,
"file_title": file_title,
"file_link": file_link,
})
Return temporary list() data that will be used later in citation extraction:
return organic_results_data
Scrape Google Scholar Cite Results using Pagination
In this section we'll use returned data from organic results and pass result_id to search query in order to extract cite results.
If you already have a list of result id's, you can skip organic results extraction:
# if you already have a list of result id's
result_ids = ["FDc6HiktlqEJ"..."FDc6Hikt21J"]
for citation in result_ids:
params = {
"api_key": "API_KEY", # SerpApi API key
"engine": "google_scholar_cite", # cite results extraction
"q": citation # FDc6HiktlqEJ ... FDc6Hikt21J
}
search = GoogleSearch(params)
results = search.get_dict()
# further extraction code..
Below the Cite extraction code snippet you'll also find an step-by-step explanation on what is going on.
import os
from serpapi import GoogleSearch
from google_scholar_organic_results import organic_results
def cite_results():
print("extracting cite results..")
citation_results = []
for citation in organic_results():
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar_cite",
"q": citation["result_id"]
}
search = GoogleSearch(params)
results = search.get_dict()
print(f"Currently extracting {citation['result_id']} citation ID.")
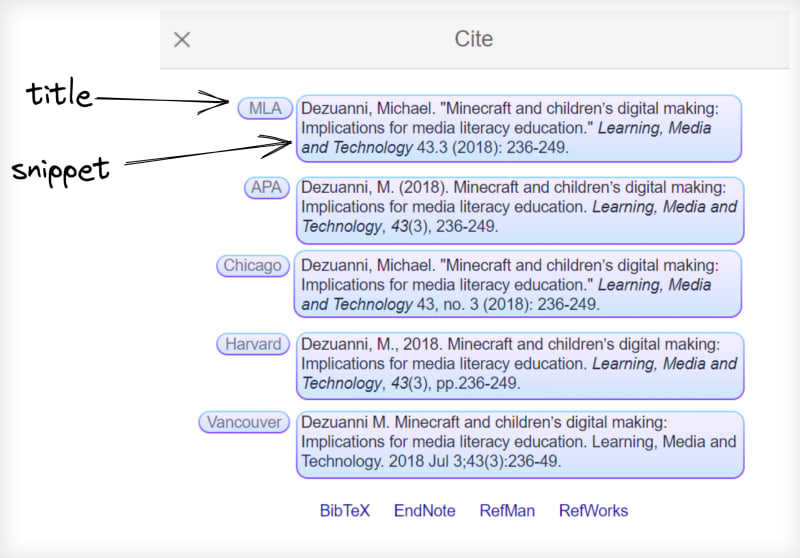
for result in results["citations"]:
cite_title = result["title"]
cite_snippet = result["snippet"]
citation_results.append({
"organic_result_title": citation["title"],
"organic_result_link": citation["link"],
"citation_title": cite_title,
"citation_snippet": cite_snippet
})
return citation_results
Explanation about Cite results extraction
Create temporary list() to store citation data:
citation_results = []
Set up a for loop to iterate over organic_results() and pass result_id to "q" search query:
for citation in organic_results():
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar_cite",
"q": citation["result_id"]
}
search = GoogleSearch(params) # from where extraction happens on the backend
results = search.get_dict() # from where JSON string is coming from
Set up a second for loop and access data as you would access a dictionary:
for result in results["citations"]:
cite_title = result["title"]
cite_snippet = result["snippet"]
Append extracted data to temporary list() as a dictionary:
citation_results.append({
"organic_result_title": citation["title"], # to know from where Cite comes
"organic_result_link": citation["link"], # to know from where Cite comes
"citation_title": cite_title,
"citation_snippet": cite_snippet
})
Return data from temporary list():
return citation_results
Save to CSV
We only need to pass returned list of dictionary from organic and cite results to DataFrame data argument and save it to_csv().
import pandas as pd
from google_scholar_organic_results import organic_results
from google_scholar_cite_results import cite_results
print("waiting for organic results to save..")
pd.DataFrame(data=organic_results()) \
.to_csv("google_scholar_organic_results.csv", encoding="utf-8", index=False)
print("waiting for cite results to save..")
pd.DataFrame(data=cite_results()) \
.to_csv("google_scholar_citation_results.csv", encoding="utf-8", index=False)
Explanation about saving results to CSV
Import organic_results() and cite_results() from where data is coming from, and pandas library:
import pandas as pd
from google_scholar_organic_results import organic_results
from google_scholar_cite_results import cite_results
Save organic results to to_csv():
pd.DataFrame(data=organic_results()) \
.to_csv("google_scholar_organic_results.csv", encoding="utf-8", index=False)
Save citation results to to_csv():
pd.DataFrame(data=cite_results()) \
.to_csv("google_scholar_citation_results.csv", encoding="utf-8", index=False)
-
dataargument insideDataFrameis your data. -
encoding='utf-8'argument just to make sure everything will be saved correctly. I used it explicitly even thought it's a default value. -
index=Falseargument to drop defaultpandasrow numbers.
Save to SQLite
By the end of this section you'll know how:
- SQLite database operates,
- save data to SQLite using
pandas, - connect and close connection to SQLite database,
- create and delete tables/columns,
- add data in a
forloop.
An example of how SQLite operates:
1. connection open
2. transaction started
3. statement executes
4. transaction done
5. connection closed
Save data to SQLite using pandas
# https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_sql.html
import sqlite3
import pandas as pd
from google_scholar_organic_results import organic_results
from google_scholar_cite_results import cite_results
conn = sqlite3.connect("google_scholar_results.db")
# save organic results to SQLite
pd.DataFrame(organic_results()).to_sql(name="google_scholar_organic_results",
con=conn,
if_exists="append",
index=False)
# save cite results to SQLite
pd.DataFrame(cite_results()).to_sql(name="google_scholar_cite_results",
con=conn,
if_exists="append",
index=False)
conn.commit()
conn.close()
-
nameis a name of SQL table. -
conis a connection to database. -
if_existswill tellpandashow to behave if the table already exists. By default, it will"fail"andraiseaValueError. In this casepandaswill append data. -
indexis to removeDataFrameindex column.
Another way of saving data using manual SQLite queries
import sqlite3
conn = sqlite3.connect("google_scholar_results.db")
conn.execute("""CREATE TABLE google_scholar_organic_results (
page_number integer,
position integer,
result_type text,
title text,
link text,
snippet text,
result_id text,
publication_info_summary text,
cited_by_count integer,
cited_by_link text,
cited_by_id text,
total_versions integer,
all_versions_link text,
all_versions_id text,
file_format text,
file_title text,
file_link text)""")
conn.commit()
conn.execute("""CREATE TABLE google_scholar_cite_results (
organic_results_title text,
organic_results_link text,
citation_title text,
citation_link text)""")
conn.commit()
# store Organic data to database
for item in organic_results():
conn.execute("""INSERT INTO google_scholar_organic_results
VALUES (:page_number,
:position,
:result_type,
:title,
:link,
:snippet,
:result_id,
:publication_info_summary,
:cited_by_count,
:cited_by_link,
:cited_by_id,
:total_versions,
:all_versions_link,
:all_versions_id,
:file_format,
:file_title,
:file_link)""",
{"page_number": item["page_number"],
"position": item["position"],
"result_type": item["type"],
"title": item["title"],
"link": item["link"],
"snippet": item["snippet"],
"result_id": item["result_id"],
"publication_info_summary": item["publication_info_summary"],
"cited_by_count": item["cited_by_count"],
"cited_by_link": item["cited_by_link"],
"cited_by_id": item["cited_by_id"],
"total_versions": item["total_versions"],
"all_versions_link": item["all_versions_link"],
"all_versions_id": item["all_versions_id"],
"file_format": item["file_format"],
"file_title": item["file_title"],
"file_link": item["file_link"]})
conn.commit()
# store Cite data to database
for cite_result in cite_results():
conn.execute("""INSERT INTO google_scholar_cite_results
VALUES (:organic_result_title,
:organic_result_link,
:citation_title,
:citation_snippet)""",
{"organic_result_title": cite_result["organic_result_title"],
"organic_result_link": cite_result["organic_result_link"],
"citation_title": cite_result["citation_title"],
"citation_snippet": cite_result["citation_snippet"]})
conn.commit()
conn.close() # explicit is better than implicit.
Explanation about saving results using manual SQLite queries
Import sqlite3 library:
import sqlite3
Connect to existing database or give it a name, and it will be created:
conn = sqlite3.connect("google_scholar_results.db")
Create Organic results table and commit changes:
conn.execute("""CREATE TABLE google_scholar_organic_results (
page_number integer,
position integer,
result_type text,
title text,
link text,
snippet text,
result_id text,
publication_info_summary text,
cited_by_count integer,
cited_by_link text,
cited_by_id text,
total_versions integer,
all_versions_link text,
all_versions_id text,
file_format text,
file_title text,
file_link text)""")
conn.commit()
Create Cite results table and commit changes:
conn.execute("""CREATE TABLE google_scholar_cite_results (
organic_results_title text,
organic_results_link text,
citation_title text,
citation_link text)""")
conn.commit()
Add extracted data to Organic table in a loop:
for item in organic_results():
conn.execute("""INSERT INTO google_scholar_organic_results
VALUES (:page_number,
:position,
:result_type,
:title,
:link,
:snippet,
:result_id,
:publication_info_summary,
:cited_by_count,
:cited_by_link,
:cited_by_id,
:total_versions,
:all_versions_link,
:all_versions_id,
:file_format,
:file_title,
:file_link)""",
{"page_number": item["page_number"],
"position": item["position"],
"result_type": item["type"],
"title": item["title"],
"link": item["link"],
"snippet": item["snippet"],
"result_id": item["result_id"],
"publication_info_summary": item["publication_info_summary"],
"cited_by_count": item["cited_by_count"],
"cited_by_link": item["cited_by_link"],
"cited_by_id": item["cited_by_id"],
"total_versions": item["total_versions"],
"all_versions_link": item["all_versions_link"],
"all_versions_id": item["all_versions_id"],
"file_format": item["file_format"],
"file_title": item["file_title"],
"file_link": item["file_link"]})
conn.commit()
Add extracted data to Cite table in a loop:
for cite_result in cite_results():
conn.execute("""INSERT INTO google_scholar_cite_results
VALUES (:organic_result_title,
:organic_result_link,
:citation_title,
:citation_snippet)""",
{"organic_result_title": cite_result["organic_result_title"],
"organic_result_link": cite_result["organic_result_link"],
"citation_title": cite_result["citation_title"],
"citation_snippet": cite_result["citation_snippet"]})
conn.commit()
Close database connection:
conn.close()
Additional useful commands:
# delete all data from the whole table
conn.execute("DELETE FROM google_scholar_organic_results")
# delete table
conn.execute("DROP TABLE google_scholar_organic_results")
# delete column
conn.execute("ALTER TABLE google_scholar_organic_results DROP COLUMN authors")
# add column
conn.execute("ALTER TABLE google_scholar_organic_results ADD COLUMN snippet text")
Full Extraction Code
import os
from serpapi import GoogleSearch
from urllib.parse import urlsplit, parse_qsl
def organic_results():
print("extracting organic results..")
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar",
"q": "minecraft redstone system structure characteristics strength", # search query
"hl": "en", # language
"as_ylo": "2017", # from 2017
"as_yhi": "2021", # to 2021
"start": "0"
}
search = GoogleSearch(params)
organic_results_data = []
loop_is_true = True
while loop_is_true:
results = search.get_dict()
print(f"Currently extracting page №{results['serpapi_pagination']['current']}..")
for result in results["organic_results"]:
position = result["position"]
title = result["title"]
publication_info_summary = result["publication_info"]["summary"]
result_id = result["result_id"]
link = result.get("link")
result_type = result.get("type")
snippet = result.get("snippet")
try:
file_title = result["resources"][0]["title"]
except: file_title = None
try:
file_link = result["resources"][0]["link"]
except: file_link = None
try:
file_format = result["resources"][0]["file_format"]
except: file_format = None
try:
cited_by_count = int(result["inline_links"]["cited_by"]["total"])
except: cited_by_count = None
cited_by_id = result.get("inline_links", {}).get("cited_by", {}).get("cites_id", {})
cited_by_link = result.get("inline_links", {}).get("cited_by", {}).get("link", {})
try:
total_versions = int(result["inline_links"]["versions"]["total"])
except: total_versions = None
all_versions_link = result.get("inline_links", {}).get("versions", {}).get("link", {})
all_versions_id = result.get("inline_links", {}).get("versions", {}).get("cluster_id", {})
organic_results_data.append({
"page_number": results["serpapi_pagination"]["current"],
"position": position + 1,
"result_type": result_type,
"title": title,
"link": link,
"result_id": result_id,
"publication_info_summary": publication_info_summary,
"snippet": snippet,
"cited_by_count": cited_by_count,
"cited_by_link": cited_by_link,
"cited_by_id": cited_by_id,
"total_versions": total_versions,
"all_versions_link": all_versions_link,
"all_versions_id": all_versions_id,
"file_format": file_format,
"file_title": file_title,
"file_link": file_link,
})
if "next" in results.get("serpapi_pagination", {}):
search.params_dict.update(dict(parse_qsl(urlsplit(results["serpapi_pagination"]["next"]).query)))
else:
loop_is_true = False
return organic_results_data
def cite_results():
print("extracting cite results..")
citation_results = []
for citation in organic_results():
params = {
"api_key": os.getenv("API_KEY"),
"engine": "google_scholar_cite",
"q": citation["result_id"]
}
search = GoogleSearch(params)
results = search.get_dict()
print(f"Currently extracting {citation['result_id']} citation ID.")
for result in results["citations"]:
cite_title = result["title"]
cite_snippet = result["snippet"]
citation_results.append({
"organic_result_title": citation["title"],
"organic_result_link": citation["link"],
"citation_title": cite_title,
"citation_snippet": cite_snippet
})
return citation_results
# example console output when extracting organic results and saving to SQL:
'''
extracting organic results..
Currently extracting page №1..
Currently extracting page №2..
Currently extracting page №3..
Currently extracting page №4..
Currently extracting page №5..
Currently extracting page №6..
Done extracting organic results.
Saved to SQL Lite database.
'''
Full Saving Code
import pandas as pd
import sqlite3
from google_scholar_organic_results import organic_results
from google_scholar_cite_results import cite_results
# One way of saving to database Pandas
print("waiting for organic results to save..")
organic_df = pd.DataFrame(data=organic_results())
organic_df.to_csv("google_scholar_organic_results.csv", encoding="utf-8", index=False)
print("waiting for cite results to save..")
cite_df = pd.DataFrame(data=cite_results())
cite_df.to_csv("google_scholar_citation_results.csv", encoding="utf-8", index=False)
# ------------------------------
# Another way of saving to database using manual SQLite queries
conn = sqlite3.connect("google_scholar_results.db")
conn.execute("""CREATE TABLE google_scholar_organic_results (
page_number integer,
position integer,
result_type text,
title text,
link text,
snippet text,
result_id text,
publication_info_summary text,
cited_by_count integer,
cited_by_link text,
cited_by_id text,
total_versions integer,
all_versions_link text,
all_versions_id text,
file_format text,
file_title text,
file_link text)""")
conn.commit()
conn.execute("""CREATE TABLE google_scholar_cite_results (
organic_results_title text,
organic_results_link text,
citation_title text,
citation_link text)""")
conn.commit()
for item in organic_results():
conn.execute("""INSERT INTO google_scholar_organic_results
VALUES (:page_number,
:position,
:result_type,
:title,
:link,
:snippet,
:result_id,
:publication_info_summary,
:cited_by_count,
:cited_by_link,
:cited_by_id,
:total_versions,
:all_versions_link,
:all_versions_id,
:file_format,
:file_title,
:file_link)""",
{"page_number": item["page_number"],
"position": item["position"],
"result_type": item["type"],
"title": item["title"],
"link": item["link"],
"snippet": item["snippet"],
"result_id": item["result_id"],
"publication_info_summary": item["publication_info_summary"],
"cited_by_count": item["cited_by_count"],
"cited_by_link": item["cited_by_link"],
"cited_by_id": item["cited_by_id"],
"total_versions": item["total_versions"],
"all_versions_link": item["all_versions_link"],
"all_versions_id": item["all_versions_id"],
"file_format": item["file_format"],
"file_title": item["file_title"],
"file_link": item["file_link"]})
conn.commit()
for cite_result in cite_results():
conn.execute("""INSERT INTO google_scholar_cite_results
VALUES (:organic_result_title,
:organic_result_link,
:citation_title,
:citation_snippet)""",
{"organic_result_title": cite_result["organic_result_title"],
"organic_result_link": cite_result["organic_result_link"],
"citation_title": cite_result["citation_title"],
"citation_snippet": cite_result["citation_snippet"]})
conn.commit()
conn.close()
print("Saved to SQL Lite database.")
# Example console output:
'''
extracting organic results..
Currently extracting page №1..
...
Currently extracting page №4..
extracting cite results..
extracting organic results..
Currently extracting page №1..
...
Currently extracting page №4..
Currently extracting 60l4wsP6Ps0J citation ID.
Currently extracting 9hkhIFu_BhAJ citation ID.
...
Saved to SQL Lite database.
'''
Links
- GitHub repository
- Extraction and CSV saving code in the Online IDE
- Google Scholar API
- SerpApi libraries
What's next
With this data it should be possible to do a research for certain discipline. A great additional feature would be to run script every week, month to get additional data. It can be done using Google Cloud Functions for example.
The follow-up blog post will be about scraping Profile results using pagination as well as Author results.
If your goal is to extract data without the need to write a parser from scratch, figure out how to bypass blocks from search engines, how to scale it or how to extract data from JavaScript - have a try SerpApi or contact SerpApi.
Add a Feature Request💫 or a Bug🐞








Top comments (0)